Research
Field-ready DNA technologies for tracing and materials, plus robust superhydrophobic surfaces for icing control.
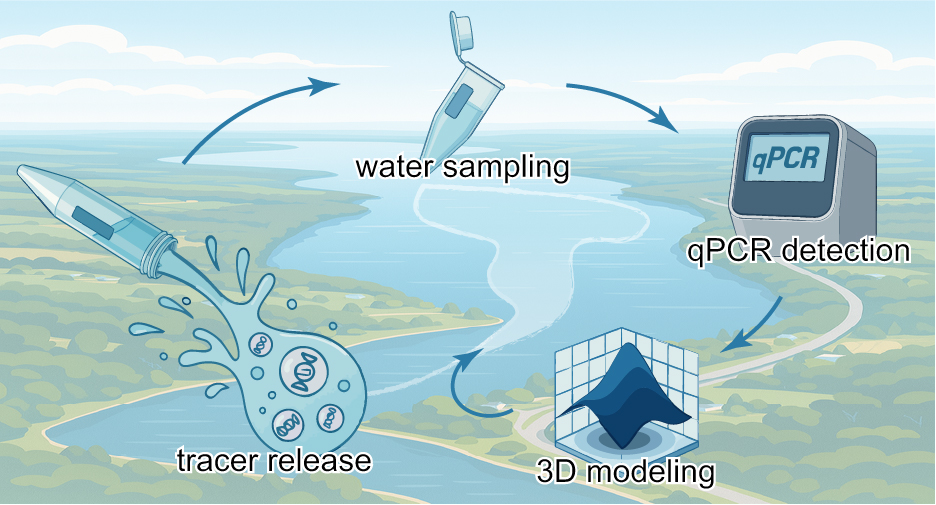
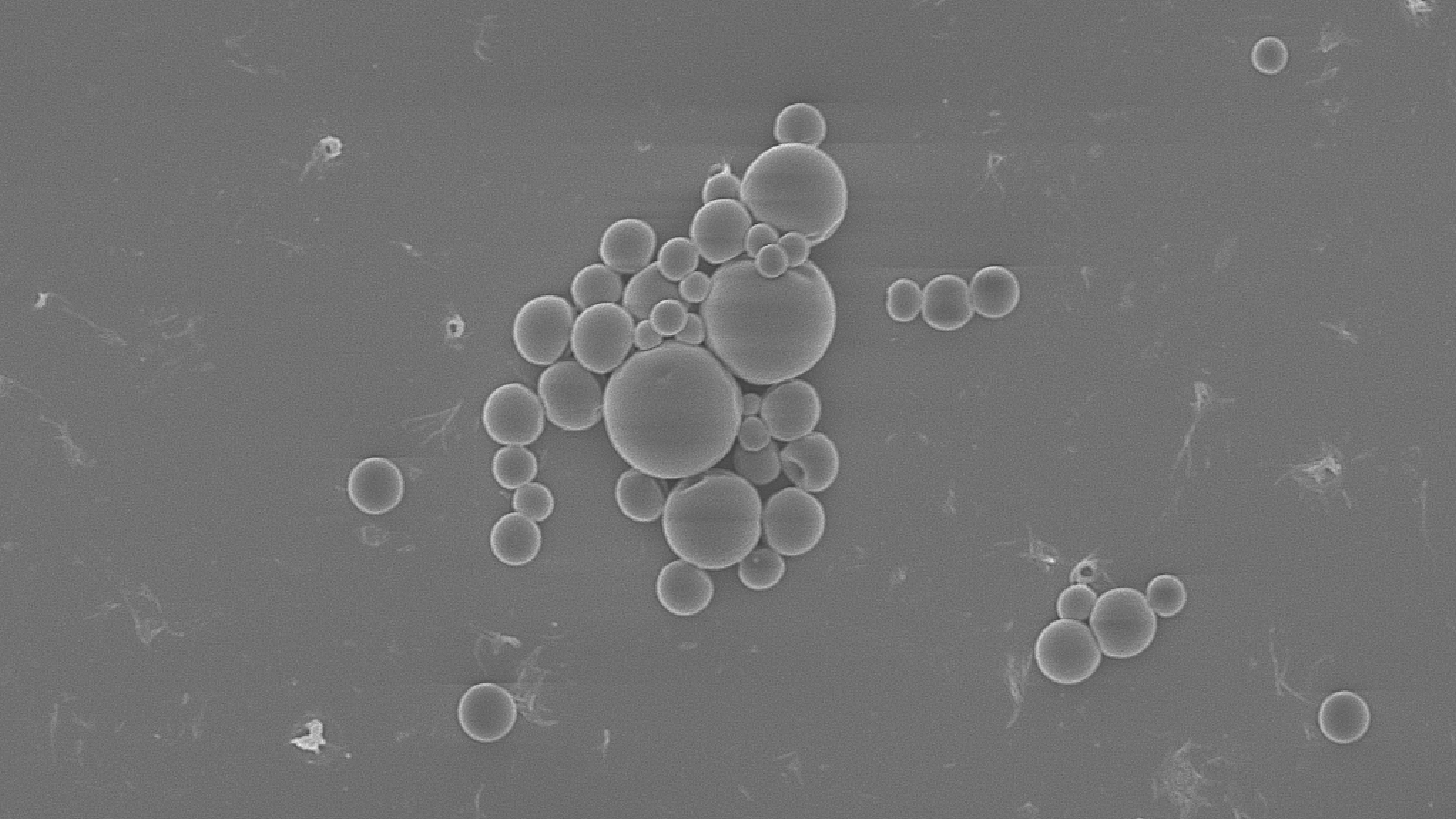
DNA-Barcoded PLGA Microspheres for Transport Mapping
DNA-barcoded PLGA microspheres mapped lake flows, detecting tracers ~7 km across ~11 km² with milligram DNA input, matching a calibrated 3D model for source attribution.
Click for Full Details
Problem
Large water bodies like lakes pose a challenge for environmental monitoring because it is difficult to track where water (and the substances in water) travels over time. Traditional tracers (dyes, salts, etc.) require prohibitively large quantities and still don't allow precise source tracking. We faced the need for a sensitive, environmental-friendly tracer that could map water flow paths across kilometers and attribute detections to specific release sources in a complex lake system.
Approach
We developed a synthetic eDNA tracer by encapsulating unique DNA barcodes in biodegradable PLGA microspheres. These particles were released into a lake and tracked via quantitative PCR (qPCR) detection of their DNA tags, while a 3D hydrodynamic model was used in parallel to map dispersion patterns and infer source locations. This approach combined polymer engineering (for particle design), molecular biology (DNA barcoding and qPCR), and environmental modeling.
Results
We executed a lake-scale field trial on Cayuga Lake, releasing uniquely barcoded DNA-PLGA microspheres and sampling 278 points over ~11 km² for ~33 hours. qPCR detected tracers to ~7 km from release with milligram-scale DNA input, and dispersion patterns aligned with a calibrated 3D hydrodynamic/Lagrangian model. The deployment validated multiplexable barcoding in one experiment and produced a robust dataset suitable for model tuning and source-attribution mapping. Outcomes to date include publication in Environmental Science & Technology, program support from DoD SERDP, and seminars delivered at ACS, UIUC, Penn State, and Cornell, underscoring both technical rigor and community interest.
Impact
This work provided one of the first lake-scale demonstration that precise spatiotemporal source tracking of eDNA is achievable. By using synthetic DNA particles, the team established a robust, standardized benchmark for eDNA transport studies. The result is a transferable tracer platform tunable by particle size and assay design for sensitive, multiplexed monitoring of water flows and contaminants. Utilities and environmental agencies could deploy such DNA tracers to map pollution sources or verify water circulation patterns with unprecedented sensitivity.
Next Steps
Ongoing work is shifting to cheap, biomass-derived DNA tracers to enable 10⁴–10⁶ times higher tracer quantities while maintaining the similar cost, and direct field detection without PCR amplification. In parallel, a portable detection device is being designed for rapid on-site readout, and new field deployments of biomass DNA tracers are planned in diverse environments to further validate and refine the approach.
Key Outcomes
- ✓ First-author paper published on ES&T (Top 5% Journal in Environmental Sciences)
- ✓ Field experiment: 11 km², ~33 h
- ✓ qPCR detection to ~7 km with < 1 mg DNA release
- ✓ Supported by DoD SERDP
- ✓ Seminars at ACS, UIUC, Penn State, and Cornell
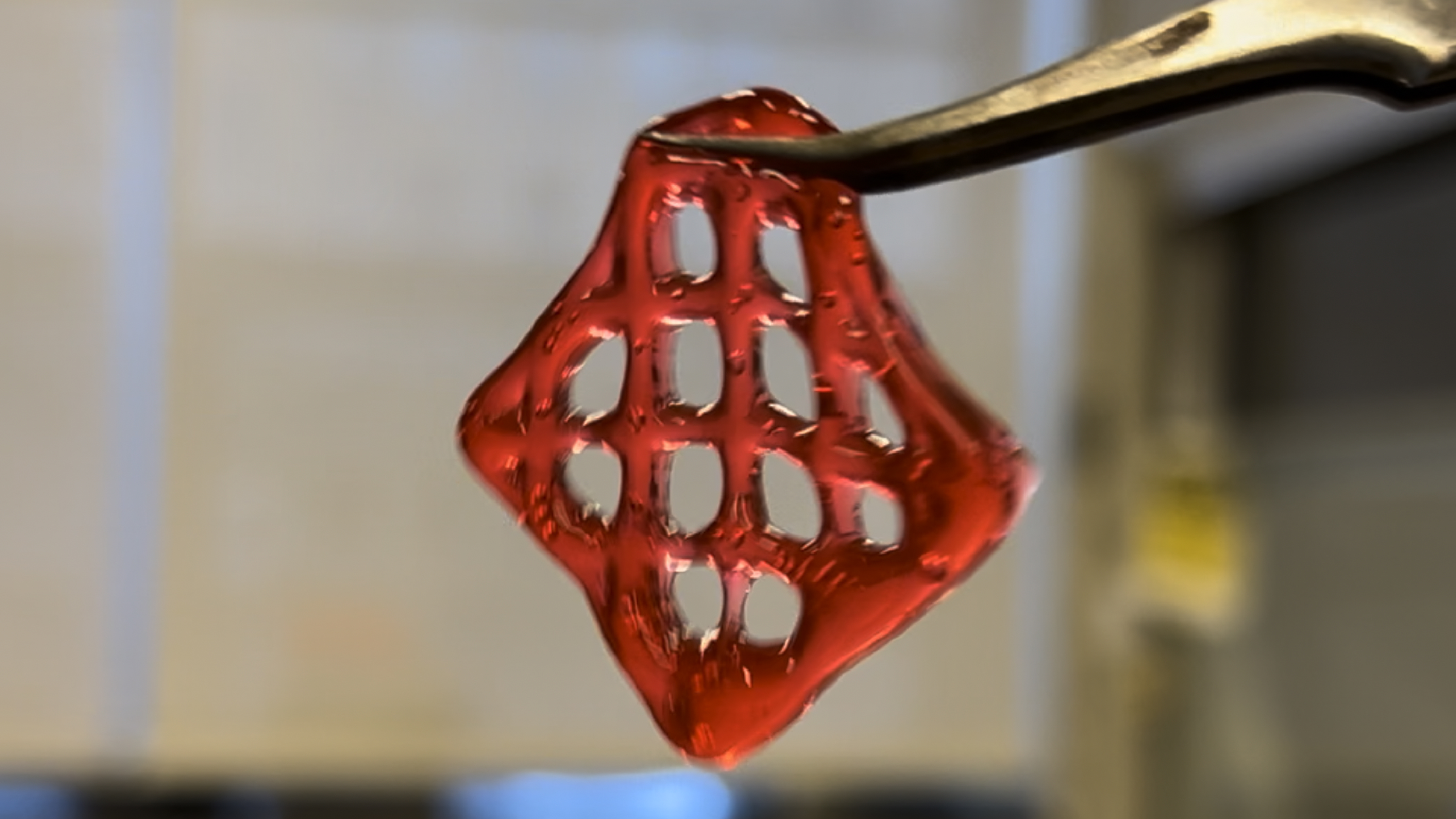
DNA Hydrogels and Composites Materials
Biomass DNA hydrogels with thermal, UV, and ionic crosslinks print and enable refillable, self-repair in architected parts, positioning DNA as a materials-grade, water-processable feedstock.
Click for Full Details
Problem
Bio-derived polymers like cellulose, starch, and collagen serve large markets, yet manufacturers face narrow processing windows, plasticizers that weaken parts, and limited chemical tunability. We address these gaps with biomass DNA as a polymer feedstock: an abundant, water-processable biopolymer with thousands of functional sites per molecule and mature biology toolkits for powerful, precise modification.
Approach
We built a biomass-DNA hydrogel platform from salmon and herring DNA and tuned three crosslink modes: thermally reversible physical crosslinks (heat/cool), UV-reversible psoralen crosslinks, and rapid ionic crosslinks with multivalent cations (e.g., Al³⁺). Formulations were tuned for application-specific rheology, stiffness, and swelling.
Results
The biomass-DNA hydrogel formed self-supporting networks that healed by thermal re-annealing, could be locked and later released with psoralen under UV, and stiffened rapidly via multivalent-ion crosslinking. Formulations printed cleanly as direct-ink-writing lattices and cast as films or membranes, with stiffness, viscosity, and swelling tuned by DNA concentration and crosslink type and density. As a demo, PolyTile 4.0's embedded channels routed dehydrated DNA precursor to cracks; upon rehydration with salts the gel set in place and sealed damage, validating on-demand delivery and multi-cycle repair in an architected part.
Impact
These results position biomass DNA as a materials-grade feedstock for low-cost, water-based processing and true reprocessability, enabling refillable self-healing and printable reinforcements at practical scale. The platform integrates with vascular composites and is extensible to concrete and porous ceramics. It can carry barrier or sensing payloads for environmental modules. Public disclosure at ACADIA 2024 indicates readiness for broader prototyping and cross-disciplinary collaboration.
Next Steps
We will map the programmability of the biomass DNA hydrogel across thermal, UV psoralen, and multivalent ion crosslinking, quantifying G′, tan δ, swelling ratios, lock and release kinetics, ionic set times, and durability over cycles, with controls and repeatability. We will log results to a compact dataset to fit simple predictors of G′ from DNA wt percent and salt. A manuscript in preparation will focus on the hydrogel as a stand-alone platform and screen gel-only use cases such as barrier membranes, sensing payloads, and direct ink writing, with clear methods and benchmarks.
Key Outcomes
- ✓ Co-First author paper published on ACADIA 2024 (Premier Conference in Computational Design)
- ✓ Supported by Cornell SPROUT Awards
- ✓ Another manuscript in preparation
- ✓ 3D-printable via DIW
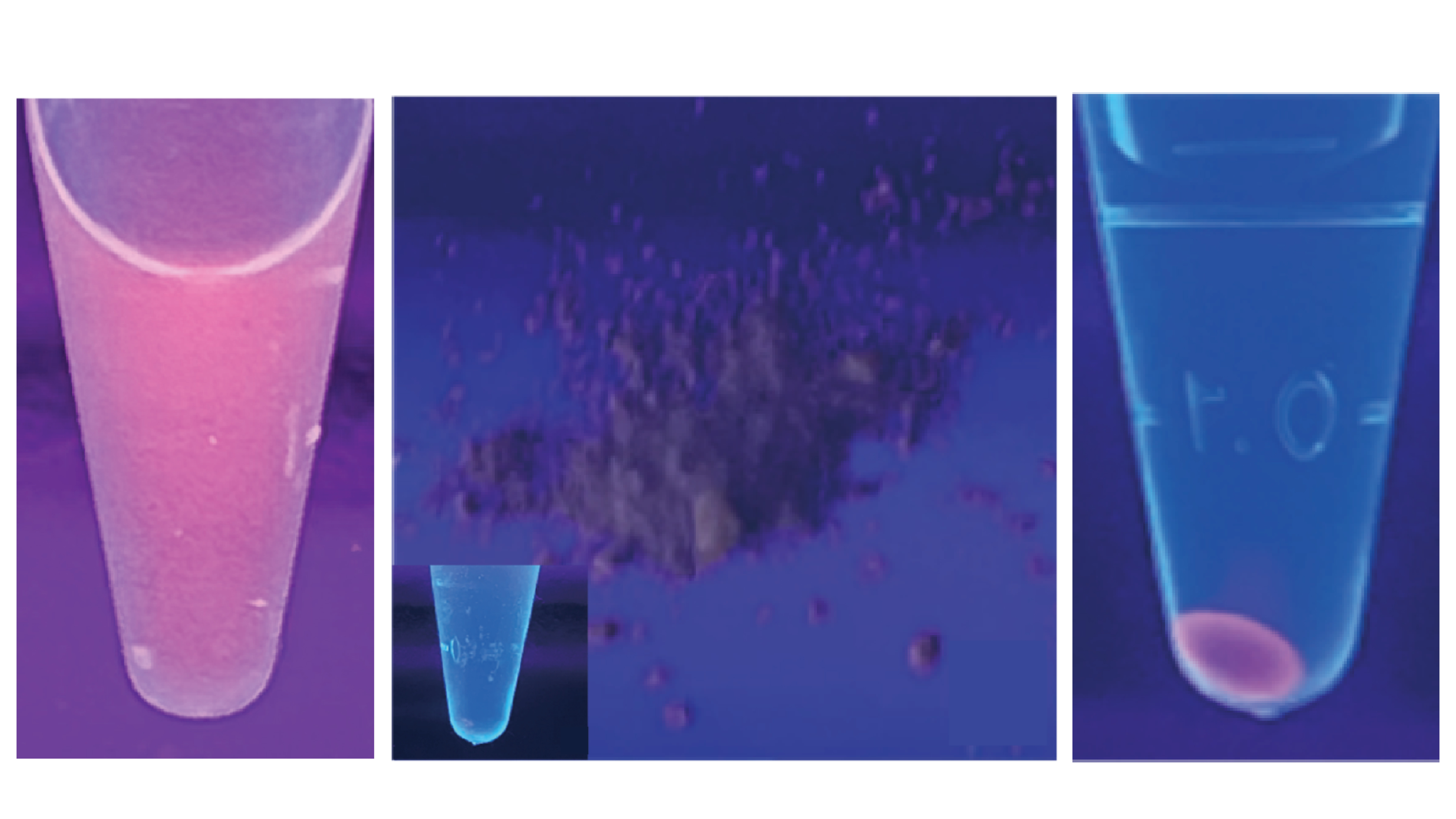
Low Cost Nucleic Acid Extraction Method Inspired by Espresso Machine
Pod-based, espresso-style extraction captured >90% DNA in 0.01–4 M salts and ~200 mg per pod, cutting ~91% using ethanol-free method toward <$1 per gram.
Click for Full Details
Problem
Bulk DNA/RNA extraction in labs still relies on manual, batch protocols with solvent-intensive precipitation. Phenol–chloroform, silica, and magnetic-bead methods consume large ethanol volumes and require repeated handling, which slows gram-scale production and raises cost.
Approach
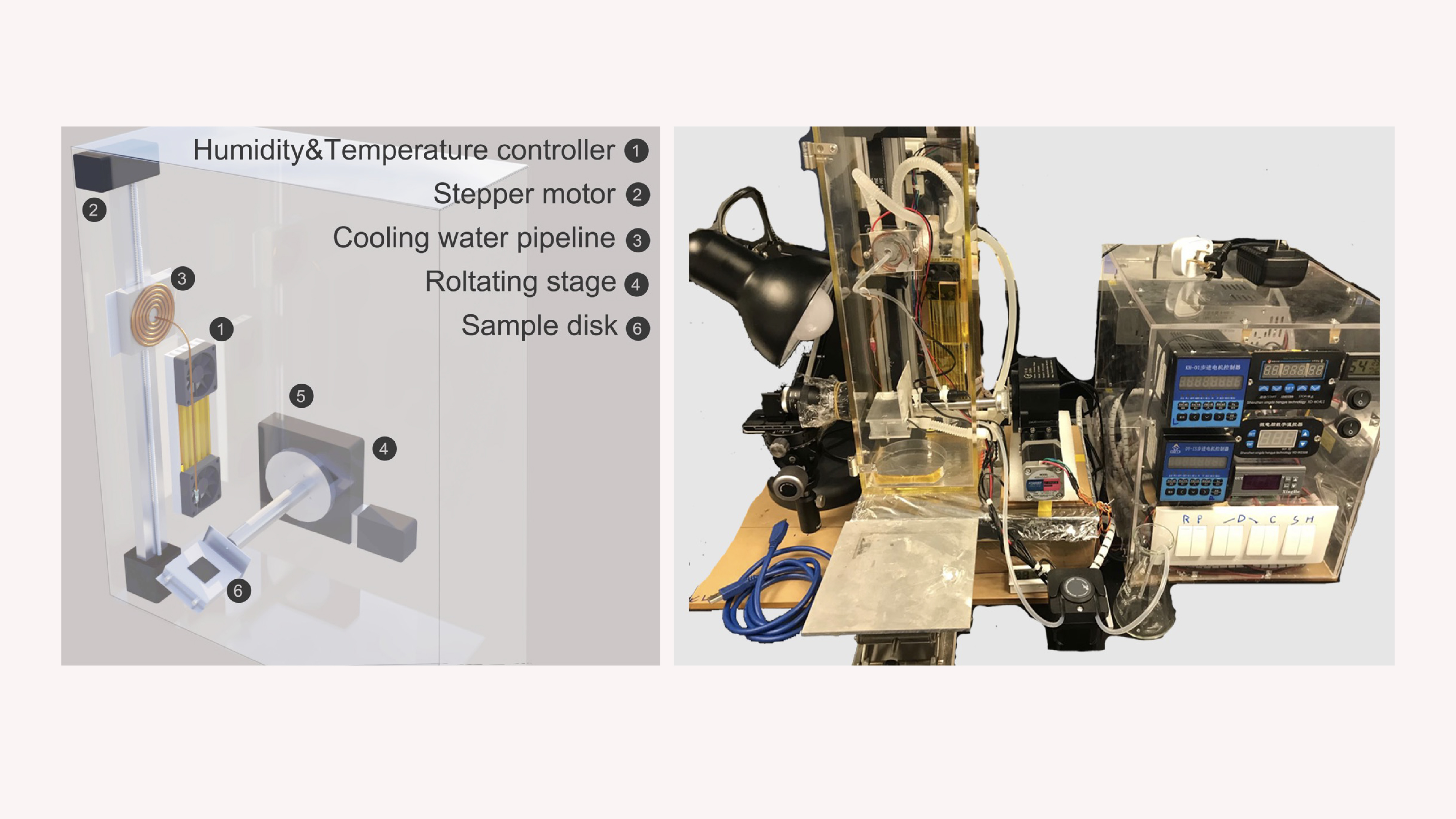
We designed an espresso-style continuous-flow extractor with a continuous inlet and outlet. Samples pass through pod cartridges packed with a custom fluoropolymer-based capture medium that adsorbs DNA/RNA in-line while permeate exits. After bind and wash, we recover nucleic acids by vapor-removing the capture medium, then recycling it for reuse. We built bench prototypes and outlined throughput scaling via pod swapping and parallelization.
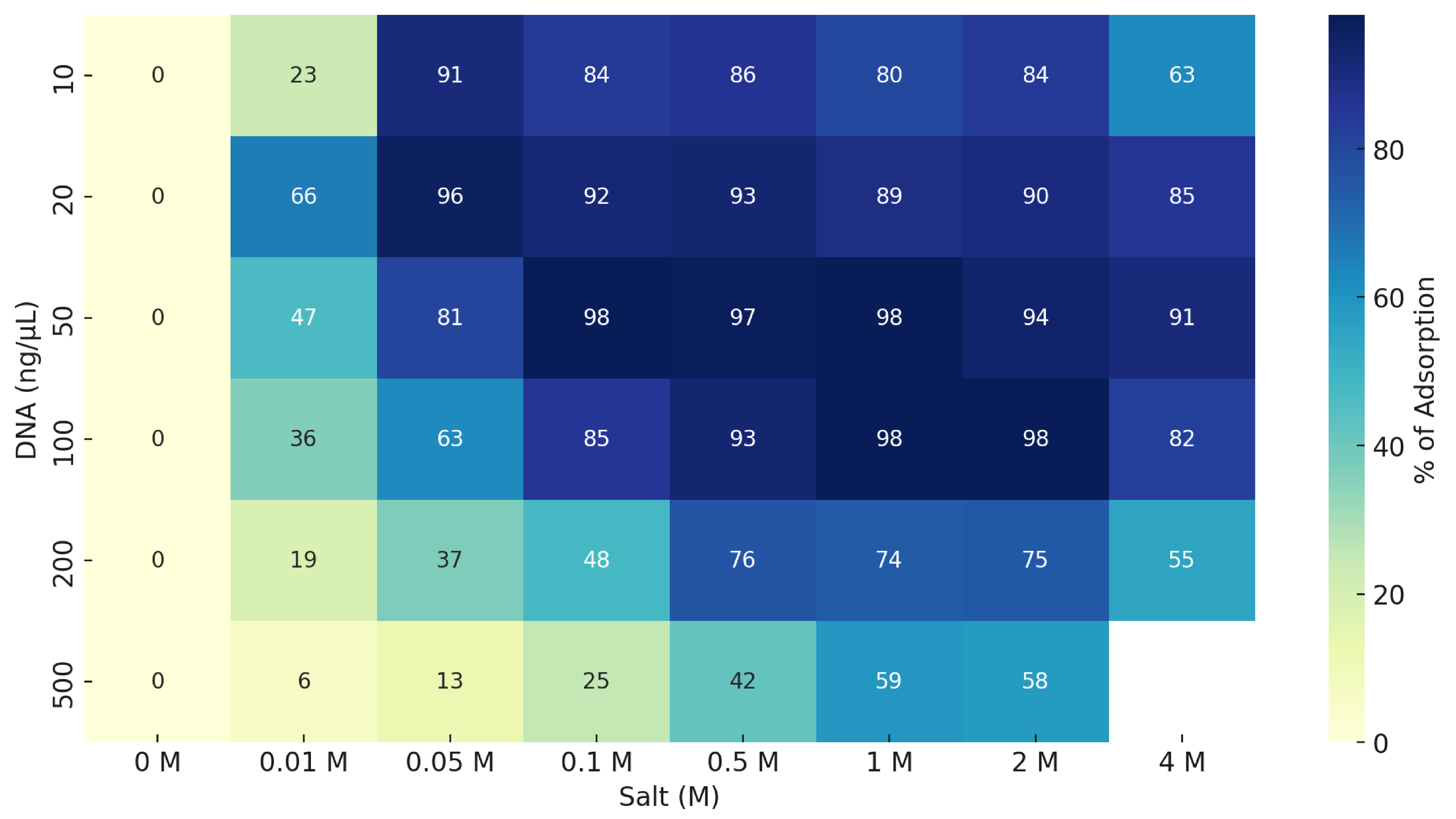
Results
Bench prototypes yielded ~200 mg DNA/RNA per pod (modeled capacity ~209 mg). Using fluoropolymer capture media, we observed >90% DNA capture across 0.01–4 M salts and 10–500 ng/µL inputs. Apparent adsorption capacity exceeded common silica media and was comparable to ion-exchange under our test conditions. Bound DNA showed reduced nuclease degradation in preliminary tests. An ethanol-free vapor recovery step cut estimated consumables by ~91% versus ethanol precipitation. Modeling indicates a path to < $1 per gram with pod reuse.
Impact
Replacing ethanol precipitation with a vapor-removable capture medium and shifting from batch to a pod-based continuous platform reduces solvent use and operator touch time. The approach targets practical gram-level DNA supply for eDNA tracers, DNA hydrogels, and routine enzymatic workflows, with a cost trajectory suitable for scale-out.
Next Steps
Finalize a continuous, automated prototype with pod parallelization and vapor-removal regeneration. Benchmark head-to-head with silica and ion-exchange across biomass sources and environmental waters, reporting recovery, purity, and nuclease resistance with replicates. Optimize capture-medium cycling, pod lifetime, and inline concentration to move toward sub-$1 per gram. Pilot on-site extraction in small field trials.
Key Outcomes
- ✓ 200 mg nucleic acid per pod
- ✓ 91% cost reduction vs ethanol precipitation
- ✓ Enzyme-friendly outputs for downstream assays
Superhydrophobic Surfaces for Icing Control and Durable Water-Repellency
Perfluoropolymer textures with electrostatic pre-charge maintained repellency after abrasion, UV, chemicals, and pressure, and cut droplet contact time ~27% at −10 °C to suppress icing.
Click for Full Details
Problem
Superhydrophobic coatings often lose the Cassie state under abrasion, chemicals, UV, or pressure. Once hysteresis rises, droplets adhere; in subzero conditions, droplets that linger beyond nucleation times freeze and anchor, degrading safety, efficiency, and maintenance budgets in aviation, energy, and infrastructure.
Approach
We combined two strategies. First, we built a controlled icing rig, pre-charged samples, and quantified impact, contact time, and rebound by high-speed imaging. Second, we patterned an all-perfluoropolymer surface into micro- and nano-textures as a monolithic layer on diverse substrates, fabricated as rigid panels, freestanding films, and peel-and-stick tapes. We then evaluated wetting, immersion stability, abrasion, chemical resistance, and UV aging.
Results
Static contact angle: 155–160°. After 1,500 abrasion cycles on 1200-grit under 100 g load, angle ≥150° with roll-off ≈10.8°; failure onset ≈1,800 cycles. No change up to 300 kPa compression; at 500 kPa, angle ≈154°, roll-off ≈11°. Doubly reentrant cavities retained ≈92.7% trapped gas after 30 days at ≈90 kPa hydrostatic pressure. Repellency persisted after 7 days in corrosive vapors, continuous 185 nm UV for 7 days, and 200 °C for 7 days. Films survived >100,000 folds; tapes endured thousands of aggressive peel cycles without delamination. At −10 °C with ≈5 µL supercooled droplets, negative pre-charge at −25 kV cut contact time from ≈15 ms to ≈11 ms (≈27% reduction) and increased rebound rate. Durability was published in The Innovation (2023); the architecture is covered by US Patent 11,839,998.
Impact
Pairing intrinsically durable, monolithic perfluoropolymer textures with an active electrostatic method that shortens droplet contact time maintains the Cassie state after real wear and suppresses icing onset. This reduces reliance on heat and chemical deicers, increases uptime for turbines and power assets, and enables practical retrofits via films and tapes aligned to certification-grade durability protocols.
Next Steps
Expand icing tests across droplet sizes, wind speeds, and substrate curvatures; report field-relevant contact-time thresholds and icing-delay statistics. Advance tape and film prototypes to qualification with standardized abrasion, UV, chemical, and pressure cycling, and evaluate electrostatic pre-charge integration for large-area, low-power operation.
Key Outcomes
- ✓ Published on The Innovation (2023) (IF 33.2, 3rd author)
- ✓ Patent filings (granted 2023)
- ✓ Best Undergraduate Thesis Award (2019)